
Data Integration & Infrastructure
Half of the lab builds the VEuPathDB ecosystem, creating ontology terms, web services, and automation that help researchers explore parasite, vector, and host datasets.
We integrate parasite genomics, bioinformatics infrastructure, and evolutionary and regulatory biology to understand how eukaryotic pathogens such as Toxoplasma gondii and Cryptosporidium evolve, regulate genes, and interact with hosts.
From global databases to parasite mitochondria, our work connects discovery science with usable infrastructure.

Half of the lab builds the VEuPathDB ecosystem, creating ontology terms, web services, and automation that help researchers explore parasite, vector, and host datasets.

We study coordinated transcription during the first 72 hours of the in vitro life cycle and search for shared regulatory motifs across apicomplexans.
Emerging genomic and functional data are mined for signatures that can improve diagnostics and point to new intervention strategies.
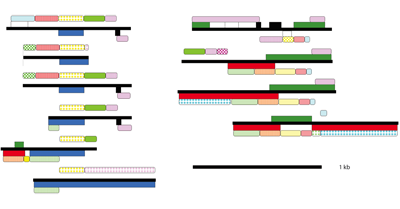
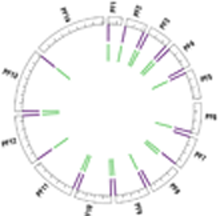
The Toxoplasma mitochondrial genome appears in many permutations, with thousands of fragments inserted into the nuclear genome, reshaping our view of organellar evolution.
From data generation to interpretation, we aim to make parasite biology discoverable and actionable.
We partner with experimental teams to generate genomes, transcriptomes, and functional datasets that anchor downstream analysis.
Multi-omics integration reveals how parasite gene regulation, genome architecture, and evolutionary history shape pathogenicity.
Our database development work ensures global researchers can visualize, query, and reuse parasite datasets with confidence.